What if a virus could reverse antibiotic resistance?
In promising experiments, phage therapy forces bacteria into a no-win dilemma that lowers their defenses against drugs they’d evolved to withstand.
- 16 September 2024
- 7 min read
- by Knowable magazine
Peering through his microscope in 1910, Franco-Canadian microbiologist Félix d'Hérelle noticed some “clear spots” in his bacterial cultures, an anomaly that turned out to be viruses preying on the bacteria. Years later, d'Hérelle would come to use these viruses, which he called bacteriophages, to treat patients plagued with dysentery after World War I.
In the decades that followed, d'Hérelle and others used this phage therapy to treat bubonic plague and other bacterial infections until the technique fell into disuse after the widespread adoption of antibiotics in the 1940s.
But now, with bacteria evolving resistance to more and more antibiotics, phage therapy is drawing a second look from researchers — sometimes with a novel twist. Instead of simply using the phages to kill bacteria directly, the new strategy aims to catch the bacteria in an evolutionary dilemma — one in which they cannot evade phages and antibiotics simultaneously.
This plan, which uses something called “phage steering,” has shown promising results in initial tests, but the scope of its usefulness remains to be proven.

There’s certainly need to find new ways to respond to bacterial infections. More than 70 percent of hospital-acquired bacterial infections in the United States are resistant to at least one type of antibiotic. And some pathogens, such as Acinetobacter, Pseudomonas, Escherichia coli and Klebsiella — classified by the World Health Organization as some of the biggest threats to human health — are resistant to multiple antibiotics. In 2019, antibacterial resistance was linked to 4.95 million deaths globally, heightening the call for more effective treatment options.
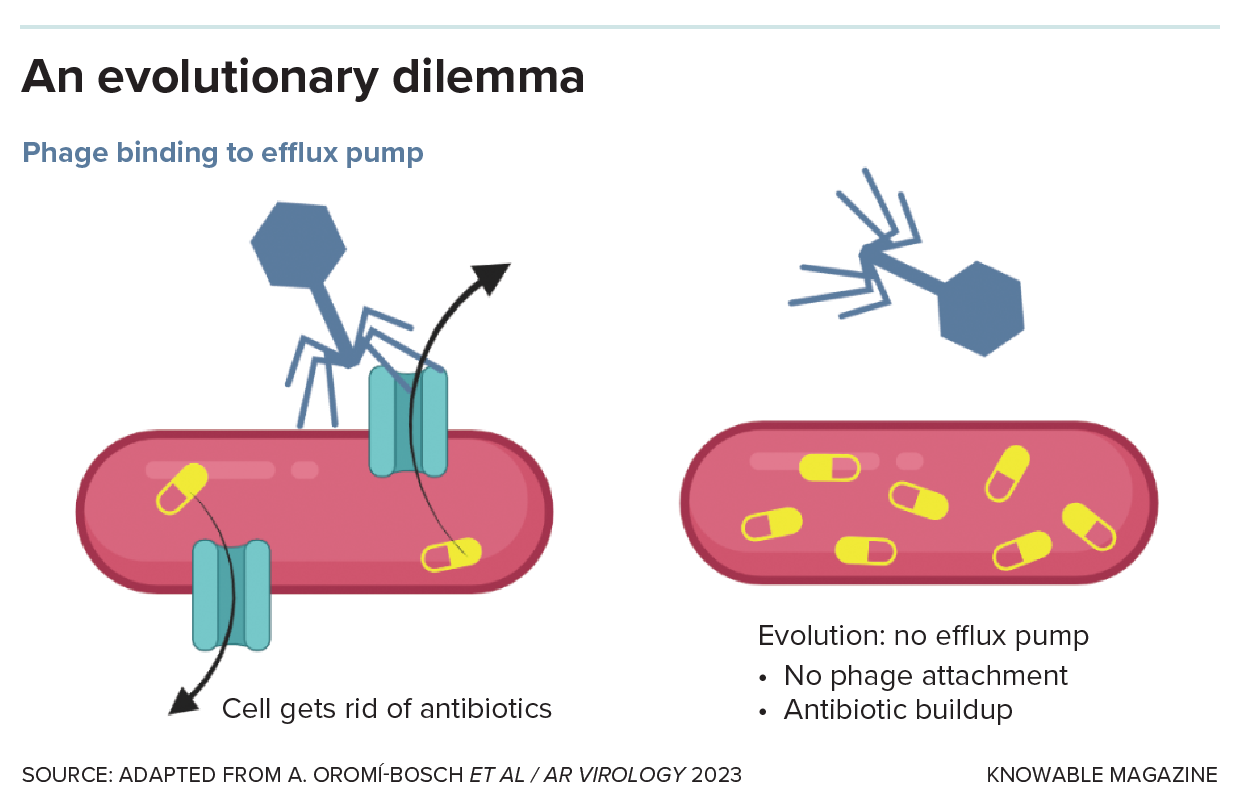
One of the ways that bacteria can evolve resistance to antibiotics is by using structures in their membranes that are designed to move unwanted molecules out of the cell. By modifying these “efflux pumps” to recognize the antibiotic, bacteria can eliminate the drug before it poisons them.
As it turns out, some phages appear to use these same efflux pumps to invade the bacterial cell. The phage presumably attaches its tail to the outer portion of the pump protein, like a key slipping into a lock, and then injects its genetic material into the cell. This lucky coincidence led Paul Turner, an evolutionary biologist at Yale University, to suggest that treating a patient with phages and antibiotics simultaneously could trap bacteria in a no-win situation: If they evolve to modify their efflux pumps so the phage can’t bind, the pumps will no longer expel antibiotics, and the bacteria will lose their resistance. But if they retain their antibiotic resistance, the phages will kill them, as Turner and colleagues explained in the 2023 Annual Review of Virology.
The result, in other words, is a two-pronged attack, says Michael Hochberg, an evolutionary biologist at the French National Centre for Scientific Research who studies how to prevent the evolution of bacterial resistance. “It’s kind of like a crisscross effect.” The same principle can target other bacterial molecules that play a dual role in resistance to viruses and antibiotics.
Turner tested this hypothesis on multidrug-resistant Pseudomonas aeruginosa, which causes dangerous infections, especially in health-care settings. This bacterium has four efflux pumps involved in antibiotic resistance, and Turner predicted that if he could find a phage that used one of the pumps as a way into the cell, the bacterium would be forced to slam the door on the phage by mutating the receptor — thereby impeding its ability to pump out antibiotics.
Sampling from the environment, Turner’s team collected 42 phage strains that infect P. aeruginosa. Out of all the phages, one, OMKO1, bound to an efflux pump, making it the perfect candidate for the experiment.
The researchers then cultured antibiotic-resistant P. aeruginosa together with OMKO1, hoping this would force the bacterium to modify its efflux pump to resist the phage. They exposed these phage-resistant bacteria, as well as their normal, phage-sensitive counterparts, to four antibiotics the bacteria had been resistant to: tetracycline, erythromycin, ciprofloxacin and ceftazidime.
As the theory predicted, the bacteria that had evolved resistance to the phage were more sensitive to the antibiotics than those that had not been exposed to the phage. This suggests that the bacteria had, indeed, been forced to lose their antibiotic resistance through their need to fight off the phage.
Other researchers have also shown that phage steering can resensitize bacteria to common antibiotics they’d become resistant to. One study, by an international research team, showed that a phage called Phab24 can be used to restore sensitivity to the antibiotic colistin in Acinetobacter baumannii, which causes life-threatening diseases.
In a second study, researchers at Monash University in Australia sampled infectious bacteria from patients. They found that several phages, including strains known as ΦFG02 and ΦCO01, were already present in some of the samples, and that A. baumannii bacteria exposed to the phages had inactivated a gene that helps create the microbe’s important outer layer, or capsule. This layer serves as the entry point for the phages, but it also helps the bacterium to form biofilms that keep out antibiotics — so removing the layer rendered A. baumannii susceptible to several antibiotics that it was previously resistant to.
In a third study, researchers from the University of Liverpool discovered that when a P. aeruginosa strain that was resistant to all antibiotics was exposed to phages, the bacterium became sensitive to two antibiotics that were otherwise considered ineffective against P. aeruginosa.
Turner’s team has used phage steering in dozens of cases of personalized therapy in clinical settings, says Benjamin Chan, a microbiologist at Yale University who works with Turner. The results, many still unpublished, have been promising so far, Chan says. Nonrespiratory infections are relatively easy to clear off, and lung infections, which the phage steering approach wouldn’t be expected to eradicate completely, often show some improvement. “I would say that we have been quite successful in using phage steering to treat difficult-to-manage infections, reducing antimicrobial resistance in many cases,” he says. But he notes that it is sometimes difficult to determine whether phage steering really was responsible for the cures.
Have you read?
Devil in the details
Phage therapy may not work for all antibiotic-resistant bacteria, says molecular biologist Graham Hatfull of the University of Pittsburgh. That’s because phages are very host-specific, and for most phages, no one knows what target they bind to on the bacterial cell surface. For phage steering to work against antibiotic resistance, the phage has to bind to a molecule that’s involved in that resistance — and it’s not clear how often that fortuitous coincidence occurs.
Jason Gill, who studies bacteriophage biology at Texas A&M University, says that it is not easy to predict if a phage will induce antibiotic sensitivity. So you always have to hunt for the right virus each time.
Gill knows from experience how complicated the approach can get. He was part of a team of researchers and doctors who used phages to treat a patient with a multidrug-resistant A. baumannii infection. Less than four days after the team administered phages intravenously and through the skin, the patient woke up from a coma and became responsive to the previously ineffective antibiotic minocycline — a striking success.
But when Gill tried a similar experiment in cell cultures, he got a different result. The A. baumannii developed resistance to the phages, but they also maintained their resistance to minocycline. “There’s not a complete mechanistic understanding,” says Gill. “The linkage between phage resistance and antibiotic sensitivity probably varies by bacterial strain, phage and antibiotic.” That means phage steering may not always work, he says.
Turner, for his part, points out another potential problem: that phages could work too well. If phage therapy kills large amounts of bacteria and deposits their remains in the bloodstream quickly, for example, this could trigger septic shock in patients. Scientists do not yet know how to address this problem.
Another concern is that doctors have less precise control over phages than antibiotics. “Phages can mutate, they can adapt, they have a genome,” says Hochberg. Safety concerns, he notes, are one factor inhibiting the routine use of phage therapy in countries like the United States, restricting it to case-by-case applications such as Turner and Chan’s.
Phage therapy may have been too high-tech for the 1940s, and even today, scientists grapple with how to use it. What we need now, says Turner, are rigorous experiments that will teach us how to make it work.
10.1146/knowable-090924-1
More from Knowable magazine
Recommended for you